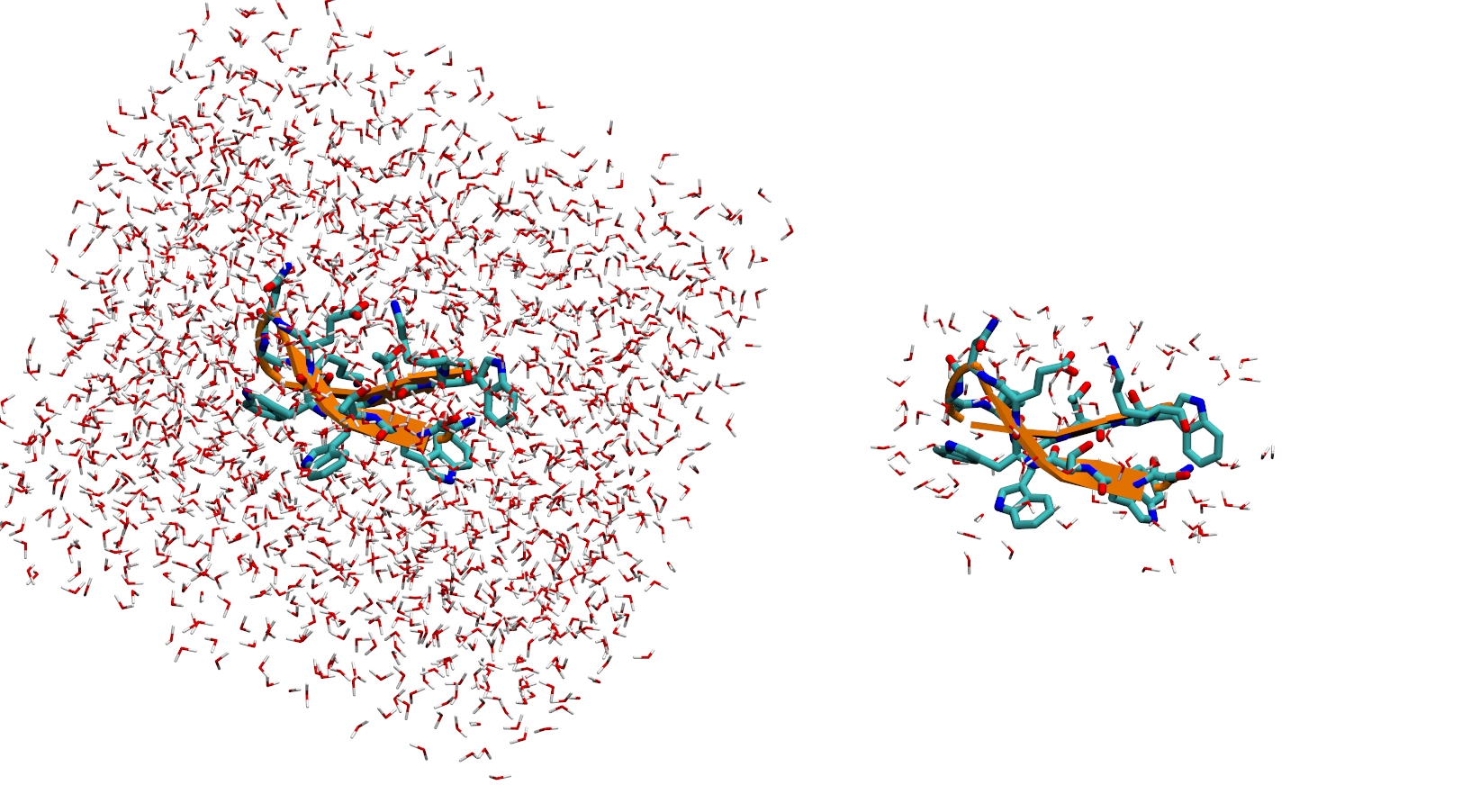
Closest solvents¶
try pytraj online:
In [7]:
# load pytraj
import pytraj as pt
# load trajectory
traj = pt.iterload('tz2.ortho.nc', 'tz2.ortho.parm7')
print(traj)
print([res.name for res in traj.top.residues if not res.is_solvent()])
In [9]:
# supposed we want to keep 100 closest solvents around protein
# specify solute mask
# and number of solvents
new_traj = pt.closest(traj, mask=':1-13', n_solvents=100, dtype='trajectory')
print(new_traj)