Parallel calculation¶
try pytraj online:
Supported
Note
pytraj does not support writing trajectory in parallel. Please check cpptraj manual for this option.
Contents
- Parallel calculation
- Example: parallel calculation with single action
- Example: parallel calculation with cpptraj’s command style
- Supported methods for
pmapif using pytraj’s methods - Supported methods for
pmapif using cpptraj’s command style - Supported methods for
openmp - Rule of thumb for choosing
pmaporopenmp? - Benchmark
- pmap API doc
Note
This is experimental design, syntax might be changed.
Example: parallel calculation with single action¶
In [1]: import pytraj as pt
In [2]: traj = pt.iterload('data/tz2.ortho.nc', 'data/tz2.ortho.parm7')
In [3]: pt.pmap(pt.radgyr, traj, n_cores=4)
Out[3]:
OrderedDict([('RoG_00000',
array([ 18.9111, 18.9365, 18.8497, 18.9045, 18.8569, 18.8892,
18.943 , 18.8888, 18.9167, 18.8707]))])
# auto-join the data
In [4]: pt.pmap(pt.radgyr, traj, n_cores=4)
��������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������Out[4]:
OrderedDict([('RoG_00000',
array([ 18.9111, 18.9365, 18.8497, 18.9045, 18.8569, 18.8892,
18.943 , 18.8888, 18.9167, 18.8707]))])
Example: parallel calculation with cpptraj’s command style¶
Rule of thumb: Only support limited cpptraj’s actions (not analysis). For now, there is no list of supported actions yet but you can perform any action in parallel if the result does not depend on the number of frame. For example, you can NOT use ‘atomicfluct’ or ‘matrix covar’, ... This parallel method is GOOD for distance, angle, rms, dihedral, vector, multidihedral, ...
# perform autoimage, then rmsfit to 1st frame
# create a reference
In [5]: ref = traj[0]
In [6]: ref
Out[6]: <Frame with 5293 atoms>
# need to explicitly do autoimage to reproduce cpptraj's serial run
# since `ref` is a single Frame, we need to provide Topology for `pt.autoimage`
In [7]: pt.autoimage(ref, top=traj.top)
��������������������������������Out[7]: <Frame with 5293 atoms>
# add to reference list
In [8]: reflist = [ref, ]
# need to explicitly specify reference by using 'refindex'
# 'refindex 0' is equal to 'reflist[0]' and so on.
# we need to explicitly specify the index so pytraj can correctly send the reference to different cores.
In [9]: pt.pmap(['autoimage', 'rms refindex 0', 'distance :3 :8', 'multidihedral phi psi resrange 3-5'], traj, ref=reflist, n_cores=4)
Out[9]:
OrderedDict([('RMSD_00001',
array([ 0. , 8.7689, 10.4369, 11.2591, 11.7381, 12.4922,
13.2369, 13.3734, 14.0668, 14.1891])),
('Dis_00002',
array([ 8.7609, 8.6396, 8.6156, 9.0299, 9.3541, 9.0833, 9.4822,
9.3856, 9.4454, 9.2076])),
('phi:3',
array([-157.0477, -153.8516, -141.437 , -168.1823, -156.3095, -152.9399,
-156.9742, -145.4146, -145.431 , -128.4447])),
('psi:3',
array([ 156.5559, 156.2962, 144.6531, 131.3405, 130.1539, 147.6546,
141.0764, 153.2045, 153.2009, 144.8442])),
('phi:4',
array([-74.4914, -79.0893, -79.84 , -57.2843, -57.6254, -93.2022,
-73.5377, -81.4212, -82.5295, -69.2517])),
('psi:4',
array([ 119.2348, 109.0394, 88.3558, 89.5826, 108.1446, 96.5808,
113.5317, 133.1828, 134.3665, 86.1773])),
('phi:5',
array([-133.9119, -138.1535, -122.0902, -121.3041, -133.1023, -124.2474,
-139.1863, -163.4636, -147.6285, -114.5396])),
('psi:5',
array([ 120.1839, 124.425 , 130.1307, 136.707 , 118.9966, 139.0724,
132.4688, 128.3897, 132.1973, 135.4395]))])
# compare to cpptraj's serial run
In [10]: state = pt.load_cpptraj_state('''
....: parm tz2.ortho.parm7
....: trajin tz2.ortho.nc
....: autoimage
....: rms
....: distance :3 :8
....: multidihedral phi psi resrange 3-5
....: ''')
....:
In [11]: state.run()
Out[11]:
CpptrajState, include:
<datasetlist: 9 datasets>
# pull output from state.data
# since cpptraj store Topology in `state.data`, we need to skip it
In [12]: state.data[1:].to_dict()
�����������������������������������������������������������Out[12]:
OrderedDict([('RMSD_00001',
array([ 0. , 8.7689, 10.4369, 11.2591, 11.7381, 12.4922,
13.2369, 13.3734, 14.0668, 14.1891])),
('Dis_00002',
array([ 8.7609, 8.6396, 8.6156, 9.0299, 9.3541, 9.0833, 9.4822,
9.3856, 9.4454, 9.2076])),
('phi:3',
array([-157.0477, -153.8516, -141.437 , -168.1823, -156.3095, -152.9399,
-156.9742, -145.4146, -145.431 , -128.4447])),
('psi:3',
array([ 156.5559, 156.2962, 144.6531, 131.3405, 130.1539, 147.6546,
141.0764, 153.2045, 153.2009, 144.8442])),
('phi:4',
array([-74.4914, -79.0893, -79.84 , -57.2843, -57.6254, -93.2022,
-73.5377, -81.4212, -82.5295, -69.2517])),
('psi:4',
array([ 119.2348, 109.0394, 88.3558, 89.5826, 108.1446, 96.5808,
113.5317, 133.1828, 134.3665, 86.1773])),
('phi:5',
array([-133.9119, -138.1535, -122.0902, -121.3041, -133.1023, -124.2474,
-139.1863, -163.4636, -147.6285, -114.5396])),
('psi:5',
array([ 120.1839, 124.425 , 130.1307, 136.707 , 118.9966, 139.0724,
132.4688, 128.3897, 132.1973, 135.4395]))])
Supported methods for pmap if using pytraj’s methods¶
In [13]: import pytraj as pt
In [14]: for method in pt.misc.parallel_info('pmap'):
....: print(method)
....:
NH_order_parameters
calc_alpha
calc_angle
calc_beta
calc_center_of_geometry
calc_center_of_mass
calc_chin
calc_chip
calc_delta
calc_dihedral
calc_distance
calc_distance_rmsd
calc_epsilon
calc_gamma
calc_mindist
calc_molsurf
calc_multidihedral
calc_nu1
calc_nu2
calc_omega
calc_phi
calc_psi
calc_radgyr
calc_rmsd_nofit
calc_rotation_matrix
calc_surf
calc_vector
calc_watershell
calc_zeta
dist
energy_decomposition
get_velocity
hbond
ired_vector_and_matrix
mean_structure
native_contacts
pucker
rmsd
rmsd_perres
volmap
Supported methods for openmp¶
In [15]: for method in pt.misc.parallel_info('openmp'):
....: print(method)
....:
calc_atomiccorr
calc_dssp
calc_pairwise_rmsd
calc_rdf
calc_watershell
closest
replicate_cell
In [16]: print("")
���������������������������������������������������������������������������������������������
Rule of thumb for choosing pmap or openmp?¶
Always try to install pytraj and cpptraj with -openmp flag.
If method supports openmp, use openmp.
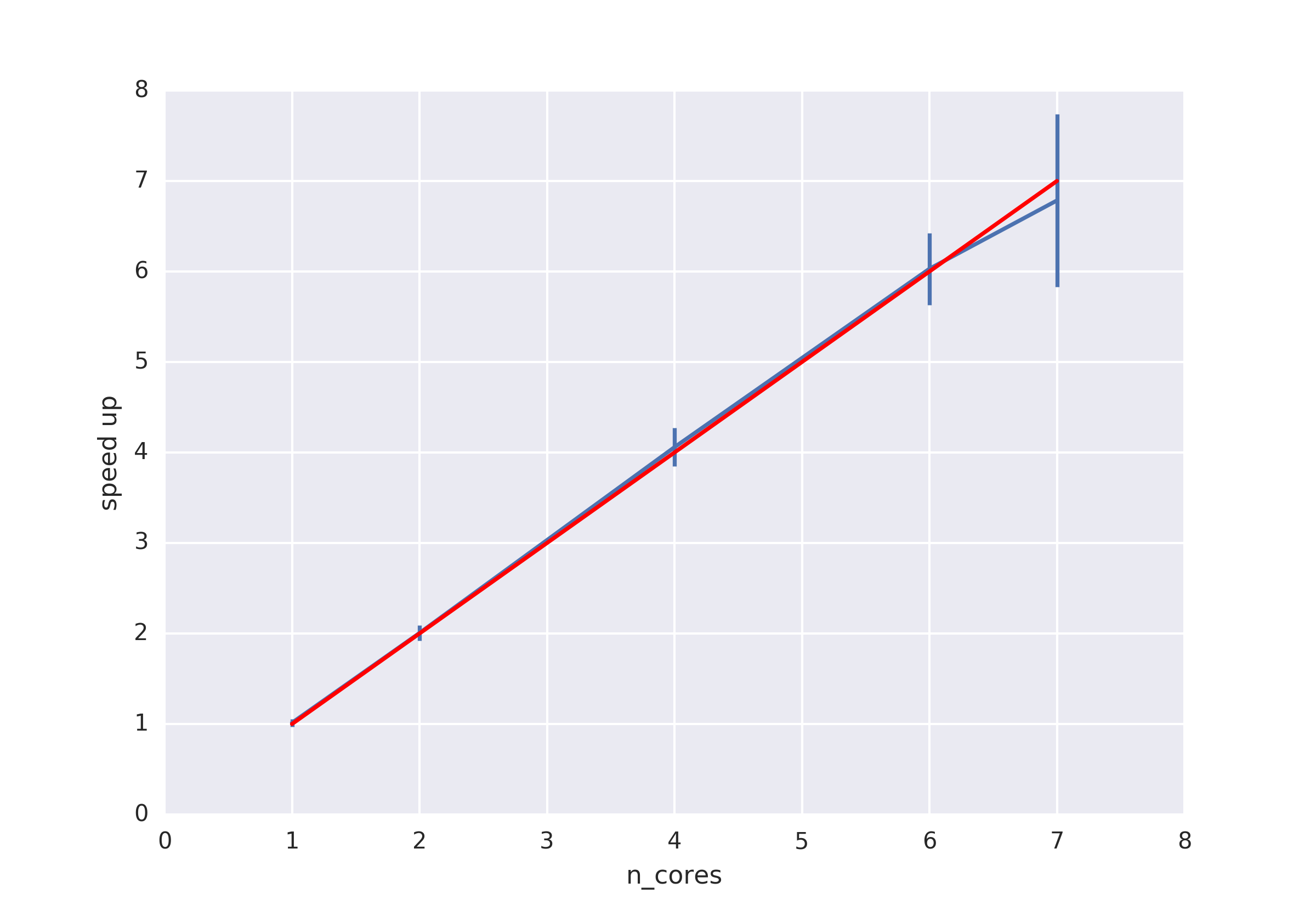
Benchmark¶
System info:
format: AMBER netcdf file
pytraj.TrajectoryIterator, 200000 frames:
Size: 58.150291 (GB)
<Topology: 13008 atoms, 4189 residues, 4174 mols, PBC with box type = truncoct>
method: pytraj.rmsd (please check the script below)
Distributed: using MPI (via mpi4py)¶
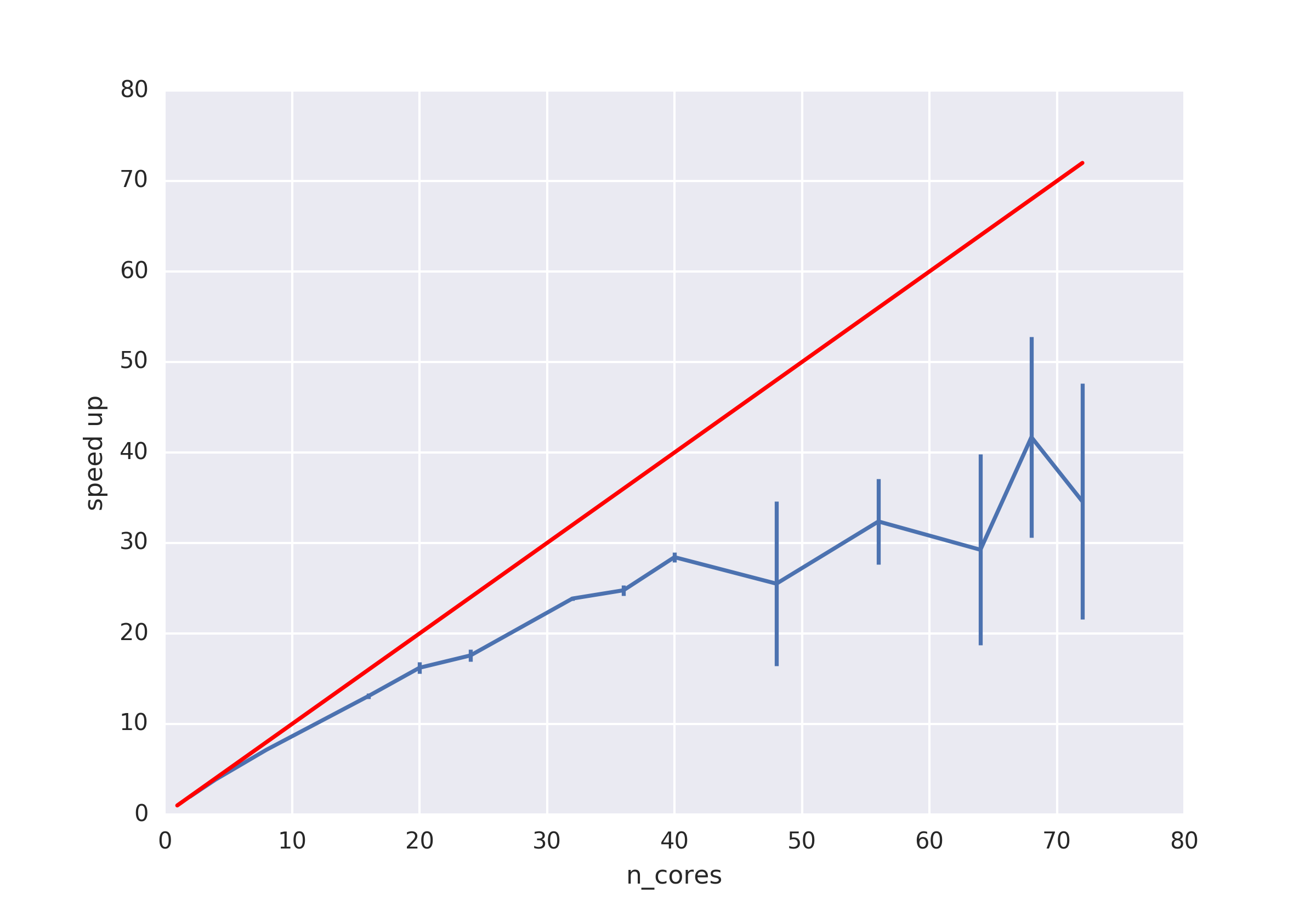
Script for multiprocessing
from multiprocessing import cpu_count
from glob import glob
import pytraj as pt
from time import time
print("max cores = {}".format(cpu_count()))
filenames = glob('mdx/md*.nc') * 10
traj = pt.iterload(filenames, 'prmtop')
print(traj)
mask = '!:WAT,Na+'
t0 = time()
pt.rmsd(traj, mask=mask)
t_serial = time() - t0
# print('serial time = ', t_serial)
func = pt.rmsd
# for n_cores in [1, 2, 4, 6, 8, 16, 20, 24]:
for n_cores in [1, 2, 4, 6, 7]:
t0 = time()
pt.pmap(func, traj, mask=mask, n_cores=n_cores, ref=traj[0])
t_par = time() - t0
print(n_cores, t_serial / t_par)
Script for MPI
fromm glob import glob
import pytraj as pt
from time import time
from mpi4py import MPI
comm = MPI.COMM_WORLD
# need to run program in serial and update serial time
serial_time = 45.
filenames = glob('mdx/md*.nc') * 10
traj = pt.iterload(filenames, 'prmtop')
mask = '!:WAT,Na+'
func = pt.rmsd
t0 = time()
x = pt.pmap_mpi(func, traj, mask=mask, ref=traj[0])
t_par = time() - t0
if comm.rank == 0:
print(serial_time/t_par)