try pytraj online:
Plot rmsd and radgyr correlation¶
In [1]:
# require: matplotlib, seaborn, pytraj
# how? "conda install matplotib seaborn"
import warnings
warnings.filterwarnings('ignore', category=DeprecationWarning)
%matplotlib inline
# add matplotlib and seaborn package
# http://matplotlib.org/
from matplotlib import pyplot as plt
# https://stanford.edu/~mwaskom/software/seaborn/index.html
import seaborn as sns
# use pytraj
import pytraj as pt
# call matrix module
from pytraj import matrix
# load trajectory from file
traj = pt.iterload('tz2.nc', 'tz2.parm7')
traj
Out[1]:
In [2]:
# caculate rmsd to 1st frame, use only backbone atoms
rmsd_data = pt.rmsd(traj, ref=0, mask='@C,N,O')
# calculate radgyr for all atoms
rg_data = pt.radgyr(traj)
print('rmsd_data', rmsd_data)
print("")
print('rg_data', rg_data)
In [3]:
# plot the correlation
# adapted from:
#sns.jointplot(rmsd_data, rg_data, kind="hex", color="#4CB391")
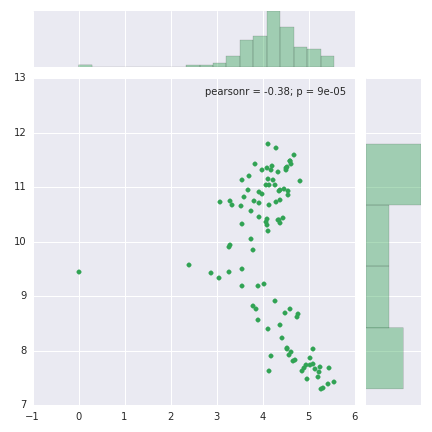
sns.jointplot(rmsd_data, rg_data, color="#31a354")
#plt.savefig('plot_rmsd_radgyr_correlation.png')
Out[3]:
(plot_rmsd_radgyr_correlation.ipynb; plot_rmsd_radgyr_correlation_evaluated.ipynb; plot_rmsd_radgyr_correlation.py)