Plot H-bond distances¶
try pytraj online:

In [1]:
import warnings
warnings.filterwarnings('ignore', category=DeprecationWarning)
# config to get better plot
%matplotlib inline
%config InlineBackend.figure_format = 'retina'
import matplotlib
matplotlib.rcParams['axes.labelcolor'] = 'green' # default green for label
matplotlib.rcParams['axes.linewidth'] = 0.5
from matplotlib import pyplot as plt
import seaborn as sb # add seaborn for pretty plot (vs default one in matplotlib)
In [2]:
# import pytraj
import pytraj as pt
# load sample data
traj = pt.iterload('tz2.nc', 'tz2.parm7')
traj
Out[2]:
In [3]:
# find hbond
hb = pt.hbond(traj)
distance_mask = hb.get_amber_mask()[0]
print('hbond distance mask: {} \n '.format(distance_mask))
angle_mask = hb.get_amber_mask()[1]
print('hbond angle mask: {} \n'.format(angle_mask))
print("hbond data")
print(hb.data) # 1: have hbond; 0: does not have hbond
In [4]:
# compute distance between donor-acceptor for ALL frames (also include frames that do not form hbond)
dist = pt.distance(traj, hb.get_amber_mask()[0])
print('all hbond distances: ', dist)
In [5]:
angle = pt.angle(traj, hb.get_amber_mask()[1])
angle
Out[5]:
Plot demo¶
In [6]:
sb.color_palette('deep', n_colors=6, desat=0.5)
sb.set_style(style='white')
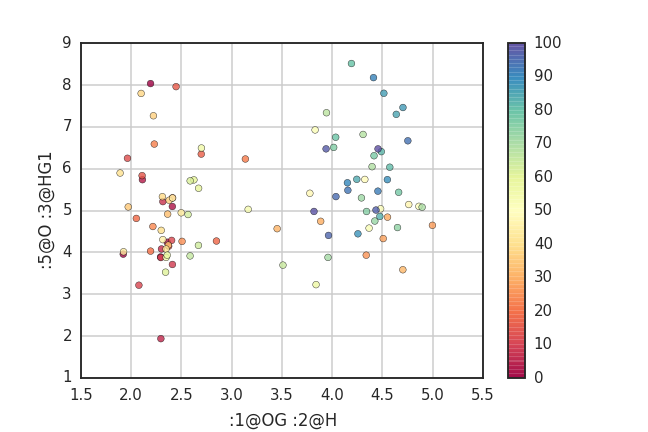
# scatter plot for distance between ':1@OG :2@H' and ':5@O :3@HG1'
# the point is colored by frame number (total frame = traj.n_frames (101))
fig = plt.scatter(dist[0], dist[1], marker='o', c=range(traj.n_frames), alpha=0.8, cmap='Spectral')
plt.colorbar()
plt.grid()
plt.xlabel(':1@OG :2@H')
plt.ylabel(':5@O :3@HG1')
Out[6]:
Stats¶
In [7]:
# Filter frames that form hbond for specific donor-acceptor
# 1st pairs: SER1_OG-TRP2_N-H
h_values = hb.data['SER1_OG-TRP2_N-H'].values # 1: For hbond; 0: not form hbond
print(h_values)
In [8]:
# ':1@OG :2@H' distance
dist[0]
Out[8]:
In [9]:
# filter distances from frames forming hbond
import numpy
h_frames = numpy.where(h_values==1)[0] # frame indices forming hbond
print('h_frames', h_frames)
In [10]:
arr = dist[0][h_frames]
print('hbond distance', arr)
In [11]:
numpy.mean(arr)
Out[11]:
In [12]:
numpy.std(arr)
Out[12]:
(plot_hbond_basic.ipynb; plot_hbond_basic_evaluated.ipynb; plot_hbond_basic.py)