RMSF¶
try pytraj online:
In [1]:
# DCD and PSF files were downloaded from
# https://github.com/MDAnalysis/mdanalysis/tree/develop/testsuite/MDAnalysisTests/data
In [2]:
import pytraj as pt
traj = pt.load('adk_dims.dcd', top='adk.psf')
traj
Out[2]:
In [13]:
# superimpose to 1st frame
pt.superpose(traj, ref=0)
# compute rmsf
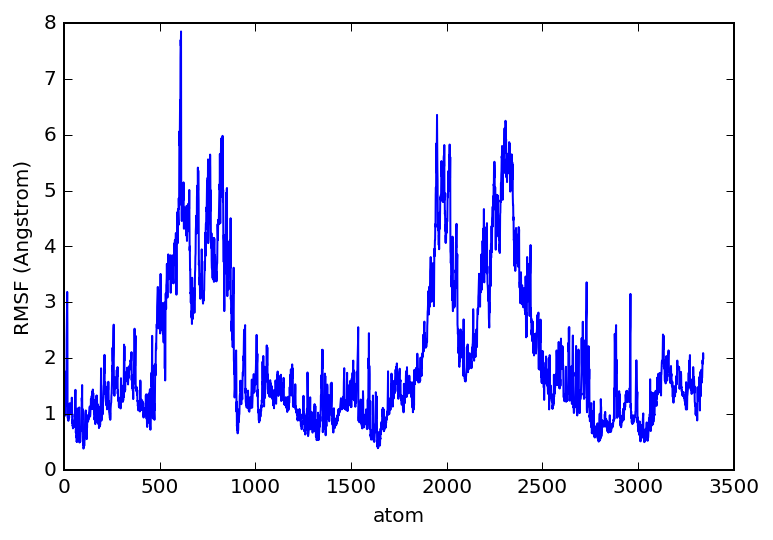
rmsf_data = pt.rmsf(traj)
rmsf_data
Out[13]:
In [14]:
# plot by using matplotlib
%matplotlib inline
import matplotlib
matplotlib.rcParams['savefig.dpi'] = 2 * matplotlib.rcParams['savefig.dpi'] # larger image
from matplotlib import pyplot as plt
plt.plot(rmsf_data.T[0], rmsf_data.T[1])
plt.xlabel('atom')
plt.ylabel('RMSF (Angstrom)')
Out[14]: